Workflows
What is a Workflow?Filters
StructuralVariants Workflow
Type: Nextflow
Creators: Laura Rodriguez-Navas, Adrián Muñoz-Civico, Daniel López-López
Submitter: Laura Rodriguez-Navas
 Tests Not available
Tests Not available
Snakemake workflow: FAIR CRCC - image conversion
A Snakemake workflow for converting whole-slide images (WSI) from the CRC Cohort ...
A prototype implementation of the Air Quality Prediction pipeline in Galaxy, using CWL tools.
ABR_Threshold_Detection
What is this?
This code can be used to automatically determine hearing thresholds from ABR hearing curves.
One of the following methods can be used for this purpose:
- neural network (NN) training,
- calibration of a self-supervised sound level regression (SLR) method
on given data sets with manually determined hearing thresholds.
Installation:
Run inside the src directory:
Installation as python package
pip install -e ./src (Installation as python
...
StructuralVariants Workflow
Type: Common Workflow Language
Creators: Laura Rodriguez-Navas, Daniel López-López
Submitter: Laura Rodriguez-Navas
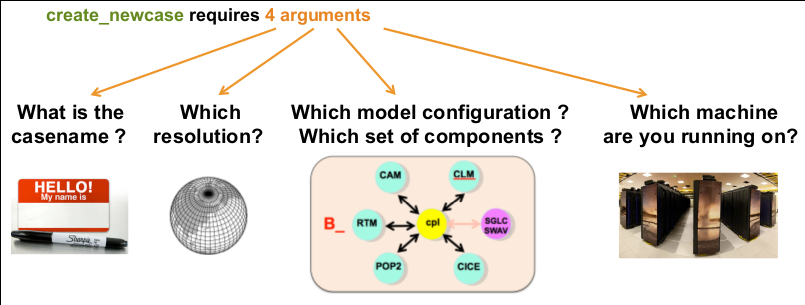
This workflow demonstrates the usage of the Community Earth System Model on Galaxy Europe.
A fully coupled B1850 compset with resolution f19_g17 is run for 1 month.
HiFi de novo genome assembly workflow
HiFi-assembly-workflow is a bioinformatics pipeline that can be used to analyse Pacbio CCS reads for de novo genome assembly using PacBio Circular Consensus Sequencing (CCS) reads. This workflow is implemented in Nextflow and has 3 major sections.
Please refer to the following documentation for detailed description of each workflow section:
A workflow for the quality assessment of mass spectrometry (MS) based proteomics analyses
ESCALIBUR
Escalibur Population Genomic Analysis Pipeline is able to explore key aspects centering the population genetics of organisms, and automates three key bioinformatic components in population genomic analysis using Workflow Definition Language (WDL: https://openwdl.org/), and customised R, Perl, Python and Unix shell scripts. Associated programs are packaged into a platform independent singularity image, for which the definition file is provided.
The workflow for analysis using Escalibur ...
